
Cyanogenic glucosides are produced in both plants and insects
The bioactive natural product class of cyanogenic glucosides is central to the center's research. This subproject focuses on understanding the genomic and evolutionary aspects of cyanogenic glucoside syntheses in various organisms.
Our work on Sorghum, Cassava, Lotus, as well as on insects, shows that the cyanogenic glucoside biosynthesis pathways in several species have evolved independently, although the enzymes may have been recruited from similar broad multigene families.
Furthermore, we have shown that in some cases the genes for cyanogenic glucoside biosynthesis are structurally organised in gene clusters so that several genes involved in making the compounds are located next to each other in the plant’s genome. Building on this work, we are now investigating putative genomic clusters in barley that may be responsible for hydroxynitrile glucoside biosynthesis. Additionally, we are assembling the transcriptome from cyanogenic ferns by next generation RNA sequencing and analyzing the cytochrome P450 encoding enzymes for signature motifs which have been identified in higher plant P450s involved in cyanogenic glucoside biosynthesis. Interestingly, CYP79s, which catalyse the first step of biosynthesis in other plants, are absent in ferns. The identification of the enzyme catalyzing the first step in cyanogenic glucoside synthesis in a fern would set the stage for further analysis of cluster plasticity over the course of evolution.
In addition to plants, our research extends to investigating the evolution of cyanogenic glucoside biosynthesis in insects. The burnet moth Zygaena filipendulae can sequester the cyanogenic glucosides lotaustralin and linamarin from their food plant, Lotus corniculatus, as well as carry out de novo biosynthesis. We will answer questions about how, when and where the cyanogenic glucoside pathway evolved in insects by phylogenetic analyses of genes cloned from species widely spread within the Zygaenoidea and Papilionoidea clades.
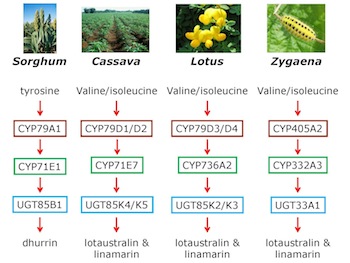
Our work on cyanogenic glucosides provides a unique entrance to resolve the question of the conditions which promote gene clustering and is therefore of fundamental importance in plant biology to understand how plasticity at the genome level is orchestrated to regulate the formation of bio-active natural products.
Principal Investigator
Elizabeth Heather Jakobsen Neilson
en@plen.ku.dk
+45 20 58 53 66
Principal Investigator

Raquel Sánchez Pérez
rasa@plen.ku.dk
+45 35 33 11 57
Principal Investigator
 Michael Lyngkjær
Michael Lyngkjær
mlyn@plen.ku.dk
+45 3353 2193
What is Plant Plasticity?

How do plants respond to threats? Obviously, plants are unable to run away let alone move when a herbivore approaches, or it's environment is stricken by drought. So how do plants cope? They have a different kind of invisible flexibility, happening in the biochemistry of their cells. Through evolution plants have acquired this special flexibility, which allow them to form an unparalleled diversity of structurally complex bioactive molecules with specialized roles. In a plethora of ways, these molecules help the plant adapt, cope, and assimilate to changes in their environment. This particular flexibility is called plant plasticity and it is the field of study for this research center. Read more about plant plasticity here.
